Welcome to Ontobat!
Ontobat is a web-based Semantic Web tool to support ontology-based biological data generation, upload, query and analysis. This is part of our efforts towards ontology-based Linked Open Data (LOD) analysis using the Sementic Web technologies. The focus of current Ontobat is on the analysis of experimental instance data generated in biomedical research. Only reliable ontologies, preferentially the ontologies in the OBO Foundry Library, will be applied in our studies.
The Ontobat components and workflow design is shown in the following figure:
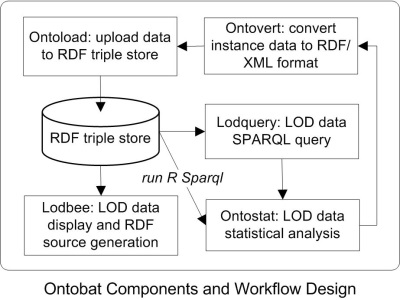
Here are some explanations of these components:
- Ontovert: Convert instance data from Excel or tab-delimited text format to ontology OWL or RDF/XML format.
- Ontoload: Automatically upload OWL-formatted instance data into a RDF triple store.
- RDF triple store: OWL-formatted instance data will be stored in a RDF triple store in a Hegroup server, or another web-accessible RDF triple store.
- Loadbee: Use Ontobee-like technology to display experimental instance data used for Ontobat data visualization.
- Lodquery: A SPARQL-based query of instance data stored in a SPARQL triple store(s).
- Ontostat: Ontology-based LOD instance data analysis.
- OntoCOG: This is an example of our Semantic Web service application. OntoCOG is designed for COG (Clusters of Orthologous Groups of proteins) enrichment analysis.
Note that as an ongoing project, some features described above have not been implemented yet. Stay tuned.
Citations:
Xiang Z, Lin Y, He Y. Ontobat: An Ontology-based semantic web approach for linked data processing and analysis. Proceedings of the 5th International Conference on Biomedical Ontologies (ICBO), Houston, Texas, USA. October 8-9, 2014. Page 93-95. [http://ceur-ws.org/Vol-1327/icbo2014_paper_58.pdf]
Cite OntoCOG: Lin Y, Xiang Z, He Y. Towards a Semantic Web application: Ontology-driven ortholog clustering analysis. International Conference on Biomedical Ontologies (ICBO), University at Buffalo, NY, July 26-30, 2011. Full length paper. Page 33 - 40.